□ The Synapse Diversity and Specificity Regulation Research Team (Director: Jaewon Ko) at DGIST (President Kunwoo Lee) reported on Tuesday the 27th that they have profiled the molecular code that constitutes brain neural circuits and discovered that it regulates specific excitatory synapse characteristics that contribute to memory of new object locations. The fine-tuning of specific excitatory synapse traits could be utilized in developing treatments for related brain developmental disorders.
□ Synapses are specialized connection sites between two neurons that constitute the basic unit of brain function. Synaptic proteins play a crucial role in transmitting neural information at these connection sites as well as in regulating neural circuits. The synaptic adhesion proteins on which this study focused are critical to brain development and function.
□ Previously, a clear understanding of how synaptic adhesion proteins bind and how that binding is regulated was lacking, and information on how these proteins operate in different parts of the brain and their impact on various brain functions was scarce.
□ Since 2011, Prof. Jaewon Ko’s Synapse Diversity and Specificity Regulation Research Team has been focusing on synaptic adhesion proteins, particularly the working principles and structure of the LAR-RPTP protein group, a pre-synaptic adhesion protein. Although previous research laid the groundwork regarding the interaction and structure of specific microexons, their functions in various of parts of the brain were not fully understood.
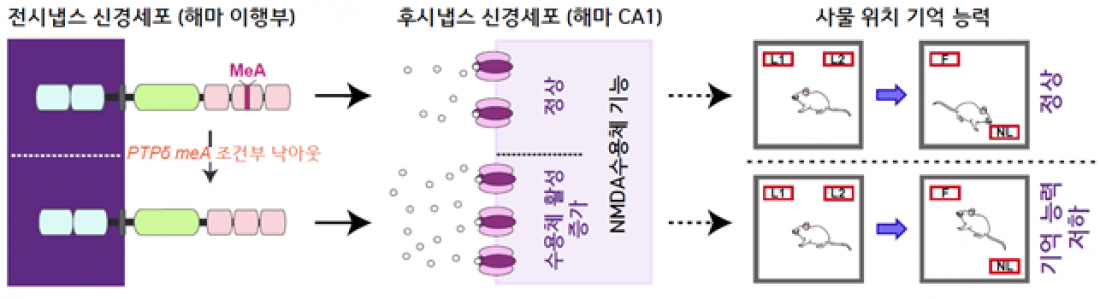
□ This study analyzed, via RNA sequencing (RNA-seq),[1] how two core microexons of the LAR-RPTP gene are expressed across different brain regions, cell types, and neural circuits. Notably, in experimental mice, a surge in the levels of specific microexons related to memory activities was observed, indicating that the synaptic adhesion pathway mediated by LAR-RPTP proteins changes while learning about new environments. This suggests the potential to regulate specific brain functions through the LAR-RPTP protein-mediated synaptic adhesion pathway.
□ Previous research has linked LAR-RPTP proteins to various brain disorders. In particular, abnormally functioning microexons in neural genes can cause brain development disorders such as autism. This study found that such brain development disorders may be related to a decline in object location memory due to specific changes in neural circuit-specific microexons. Thus, the findings could provide important clues for developing new brain disease treatments targeting various post-synaptic protein complexes associated with LAR-RPTP microexons.
□ Research Director Jaewon Ko said, "This study is the first to provide key clues on how LAR-RPTP, a critical synaptic regulator, uses microexon molecular codes to control specific synaptic traits in various neural circuit contexts. We are conducting follow-up research to elucidate the molecular mechanisms that finely regulate these microexon levels."
□ The research achievements were jointly accomplished by Prof. Kyung Ah Han of DGIST's Department of Brain Sciences and Dr. Taek Han Yoon, a graduate of the integrated master's and doctoral program in brain sciences. An online article reporting the study’s findings was published on February 22nd in the internationally renowned academic journal Nature Communications, which has an impact factor of 16.6. The study was supported by the Ministry of Science and ICT, the National Research Foundation of Korea’s Research Leader Program, the Sejong Science Fellowship, and the Daegu Gyeongbuk Institute of Science and Technology's Fostering Future-Leading Specialization Research.
- Ccorresponding Author E-mail Address : [email protected]
[1] RNA sequencing: A technique that analyzes the nucleotide sequences of all RNA transcripts transcribed from DNA in a specific sample. It can provide information about gene expression levels, nucleotide sequence variations, alternative splicing, and single nucleotide polymorphisms (SNPs).